Contact
SIRET Research Group
Department of Software Engineering
Faculty of Mathematics and Physics
Charles University
Malostranské nám. 25,
118 00 Prague
Czech Republic
| email: | info@siret.cz |
| phone: | +420 95155 4227 |
News
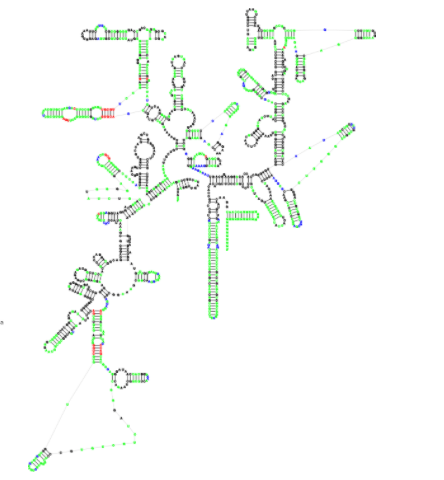
- 22.06.2021 new publication: R2DT is a framework for predicting and visualising RNA secondary structure using templates
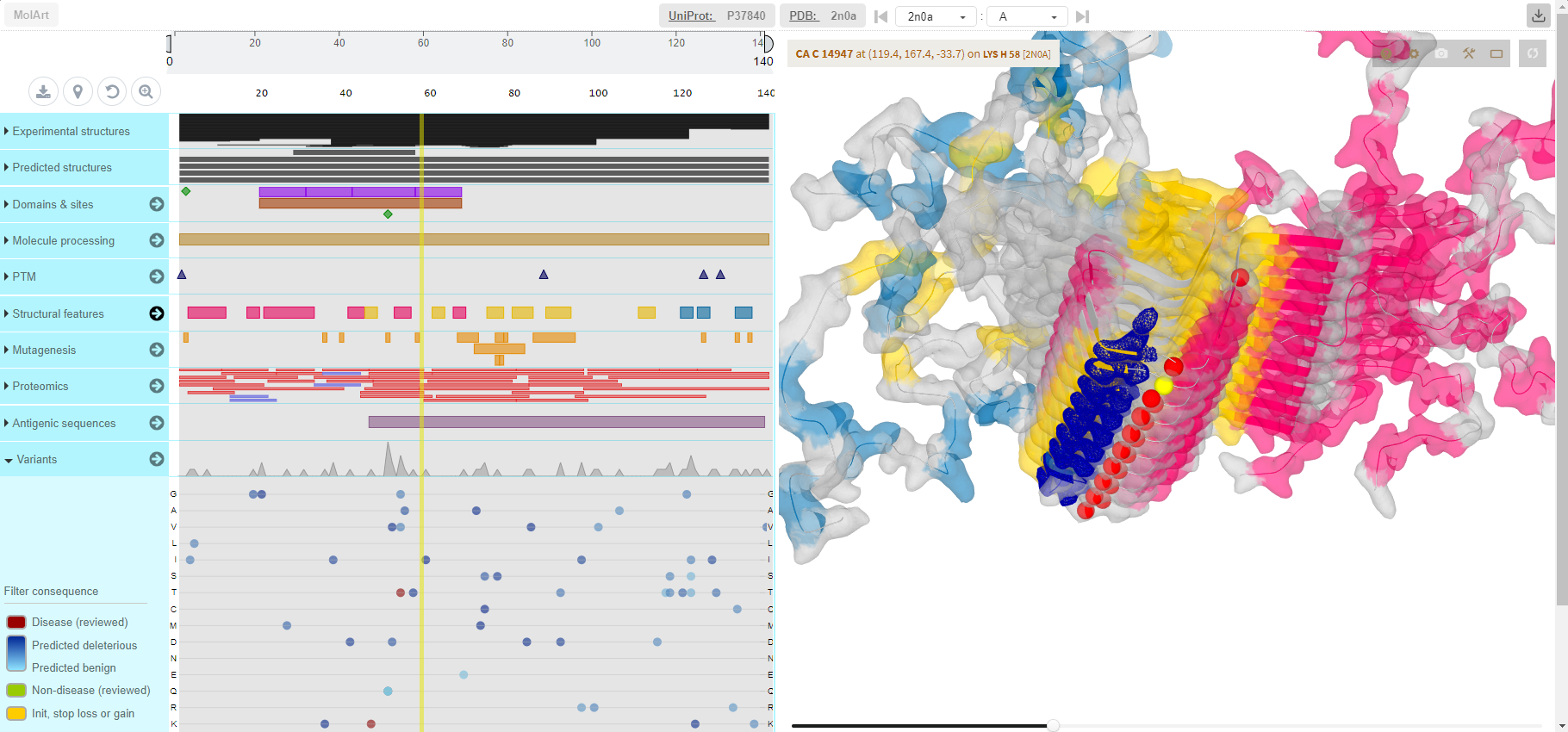
- 23.03.2021 new project: MolArt
- 23.03.2021 new project: Traveler
- 01.10.2020 new publication: RNAcentral 2021: secondary structure integration, improved sequence search, and new member databases
- 11.09.2020 new publication: Comprehensive characterization of amino acid positions in protein structures reveals molecular effect of missense variants
Copyright © 2023 SIRET Research Group. Similarity is our job.