SIRET Research Group
Department of Software Engineering
Faculty of Mathematics and Physics
Charles University
Malostranské nám. 25,
118 00 Prague
Czech Republic
| email: | info@siret.cz |
| phone: | +420 95155 4227 |
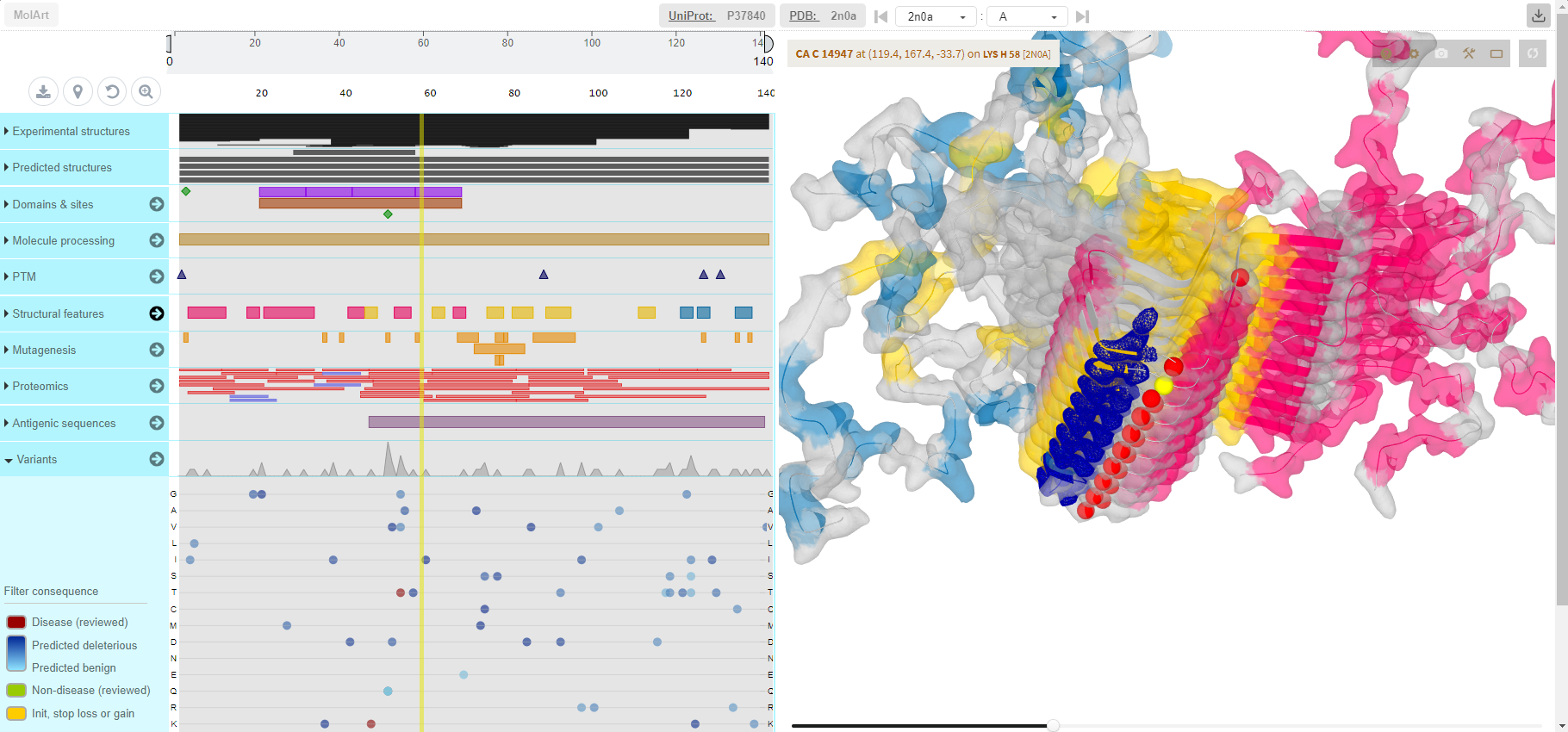 MolArt
(MolArt is a responsive, easy-to-use JavaScript plugin which enables users to view annotated protein sequence and overlay the annotations over a corresponding experimental or predicted protein structure.)
MolArt
(MolArt is a responsive, easy-to-use JavaScript plugin which enables users to view annotated protein sequence and overlay the annotations over a corresponding experimental or predicted protein structure.)
MolArt is a responsive, easy-to-use JavaScript plugin which enables users to view annotated protein sequence (including variation data from large scale studies) and overlay the annotations over a corresponding experimental or predicted protein structure. It couples protein sequence annotation capabilities provided by ProtVista (or more precisely its modified responsive version implemented when developing MolArt) with structure visualization capabilities provided by LiteMol. Since it does not have any software dependencies and all the data are obtained on the fly, it is easy to integrate it to any web page. |
 Molpher
(Software tool for exploration of the chemical space)
Molpher
(Software tool for exploration of the chemical space)
Molpher aims to be a scalable and interactive software tool to aid exploration of chemical space, the vast universe containing all possible compounds. Many areas of chemical biology, such as drug discovery, rely heavily on chemical libraries offering compounds usable in the industrial processes. Given a set of molecules with desired characteristics, Molpher explores their common neighbourhood based on structural similarity, as it represents promising part of the chemical space to find new additions into those libraries. In order to decrease the chance of missing interesting parts of the space, Molpher offers the human researcher to observe and interactively alter the exploration process. Generated subspace is expected to be further tested for synthesizability and biological activity by other software tools. Among main features, Molpher offers optimized parallel exploration algorithm, compound logging, dimension-reduced visualization of chemical space and interactive widget-based GUI. Codebase is extensible in terms of additional morphing operators, chemical fingerprints, similarity measures and visualization strategies to allow further experiments. |
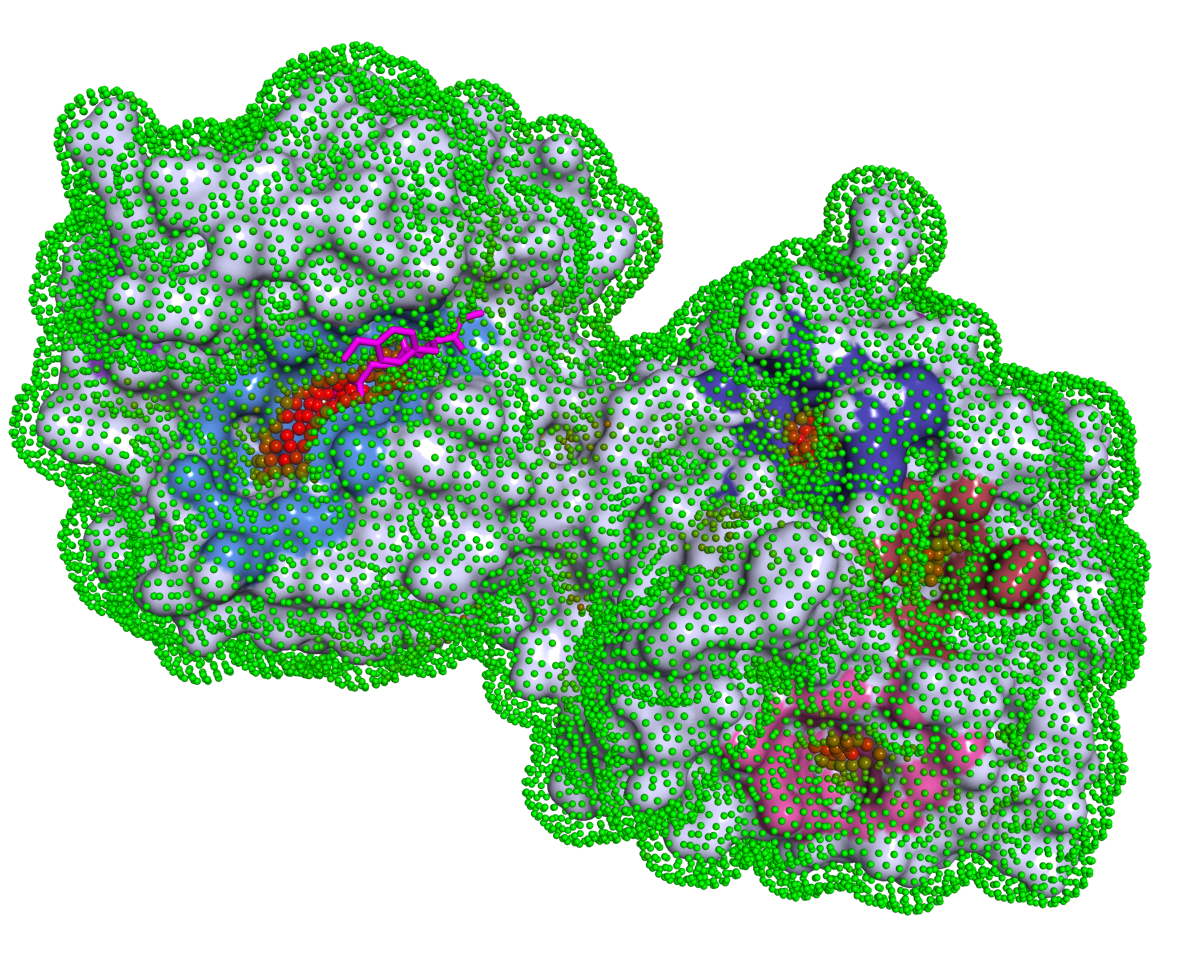 P2Rank
(Protein binding site prediction)
P2Rank
(Protein binding site prediction)
P2Rank is a machine learning based method for prediction of binding sites from protein structure. P2Rank uses Random Forests classifier to infer ligandability of local chemical neighborhoods near the protein surface which are represented by specific near-surface points and described by aggregating physico-chemical features projected on those points from neighboring protein atoms. The points with high predicted ligandability are clustered and ranked to obtain the resulting list of binding site predictions. P2Rank is freely available at https://prankweb.cz/.
|
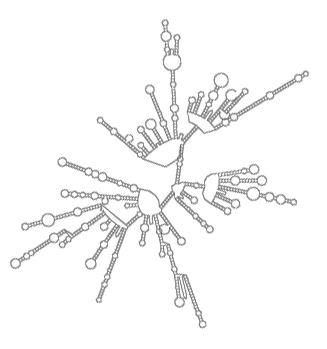 rPredictor
(Infrastructure consisting of tools, a database and web interface that together enable predicting ribosomal RNA secondary structure and analysing it.)
rPredictor
(Infrastructure consisting of tools, a database and web interface that together enable predicting ribosomal RNA secondary structure and analysing it.)
rPredictor is a bioinformatical infrastructure consisting of tools, a database and a web interface that together enable predicting ribosomal RNA secondary structure and analysing it. The predicted structures, their analyses and further details about the rRNA molecules are accessible through this website.
The aim of rPredictor is to develop and deploy a technique of predicting ribosomal RNA secondary structure and make the resulting structural information readily available. At the same time, the rPredictor database contains rich annotations of rRNA structures and the underlying sequences.
The project is being developed at the Faculty of Mathematics and Physics, Charles University in Prague in close cooperation with the bioinformatics laboratory of Microbiology Institute AVČR. |
 SETTER
(RNA structure similarity search)
SETTER
(RNA structure similarity search)
SETTER web-server utilizes SETTER (SEcondary sTructure-based TERtiary Structure Similarity Algorithm) method for fast and accurate structural pairwise alignment. The server is capable of comparing a pair of RNA structures or using one strucutre as a query and search against a user-defined database of RNA structures. The efficiency of the algorithm is given by the decomposition of the RNA structure into the set of non-overlapping generalized secondary structure motifs (GSSUs). GSSU usually resembles a hairpin motif possibly containing bulges and/or internal loops in its stem part. A segmentation to GSSUs offers good scalability with respect to the structure size (SETTER scales linearly with the structure size) because the number of residues in GSSUs (SETTER scales quadratically with the GSSU size) generally does not increase with increased size of the RNA structure. The underlying SETTER algorithm is both accurate and very fast, and does not impose limits on the size of aligned RNA structures. SETTER is able to compare a pair of even the largest RNA structure in less than one minute. |
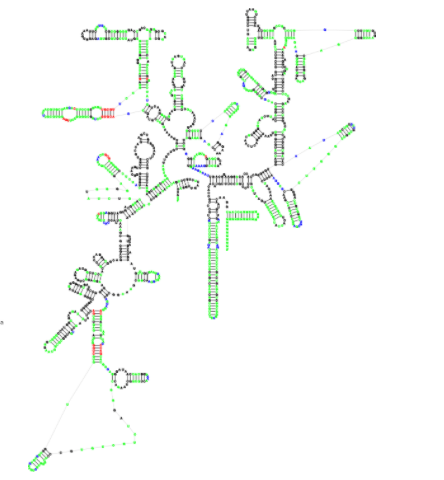 Traveler
(Traveler is an RNA sescondary structure visualization tool implementing a template-based approach enabling to lay out even the largest RNA structures in the standard orientation.)
Traveler
(Traveler is an RNA sescondary structure visualization tool implementing a template-based approach enabling to lay out even the largest RNA structures in the standard orientation.)
Visualization of RNA secondary structures is a complex task, and, especially in the case of large RNA structures where the expected layout is largely habitual, the existing visualization tools often fail to produce suitable visualizations. This led us to the idea to use existing layouts as templates for the visualization of new RNAs similarly to how templates are used in homology-based structure prediction. Traveler is a software tool enabling visualization of a target RNA secondary structure using an existing layout of a sufficiently similar RNA structure as a template. Traveler is based on an algorithm which converts the target and template structures into corresponding tree representations and utilizes tree edit distance coupled with layout modification operations to transform the template layout into the target one. Traveler thus accepts a pair of secondary structures and a template layout and outputs a layout for the target structure. Traveler is used by RNACentral for visualization of all the secondary structures available in RNACentral.
|
|
PGRTree
(Plugin for Indexing Multidimensional Data in PostgreSQL Using R-tree)
In commercial database platforms, the standard search over multiple attributes is provided by B+-tree (or it’s variants) with compound keys. On the other hand, such systems provide also multidimensional indexing, however, just for spatial purposes (such as GIS or CAD applications) and use special data types and querying syntax. Our solution allows to apply R-tree index (hence multi-dimensional indexing structure) in the same way as B+-tree with compound keys is applied. The solution is delivered as a plugin for PostgreSQL database. |
|
Exploration Portal
(Image exploration demo)
Exploration portal is a demonstration application for Multimedia exploration framework and a logical successor of SIR. The portal uses all framework features and implements example use cases for all important framework parts. The portal can be used for exploration of static data sets, Bing search results and personal Facebook albums. It provides a variety of configuration options to affect feature extraction, similarity model, index creation and many other parameters. The result is visualized using a similarity-based layout and supports different query options, such as zoom-in, zoom-out, multi-query or panning in 4 different directions. |
|
Find the image
(Online tool for comparisons of different multimedia exploration approaches)
Find the image is an artificial search scenario designed for testing and comparison of our exploration techniques. The task is to use a web-based exploration application to find as much images from a predetermined class as possible. This predetermined class should correspond to a search intention that cannot be easily transformed to a text-based query or to a query-by-example. |
|
Multimedia exploration framework
(Creation of efficient multimedia exploration applications)
Multimedia exploration framework is an extensible solution for creation of multimedia exploration applications. It uses a modular architecture and already provides several implementantions for every component. Besides managing the software architecture and data flow, the framework also takes care of data source management, data retrieval, feature extraction, distance computation, metric indexing, query execution, data visiualization and GUI creation. Contact the developers directly if you are interested in building an application using our framework. |
 SIR
(Smart image retrieval)
SIR
(Smart image retrieval)
When determining visual similarity of two images, it is evaluated on feature representations which consist of some content-based image properties. The conventional feature representations aggregate and store these properties in global feature histograms (e.g., Recent feature representations, however, adaptively aggregate local image features in more flexible feature signatures, which can be Currently, the SIR engine operates in a demo mode as a standalone image search engine. In order to manage large image collections in real time, the engine employs original database indexing technology. The SIR engine also includes meta-search functionality that allows to augment/rerank/explore results provided by other image search engines, such as Google Images and others. The actual version of the online re-ranking and exploration tool employes the particle physics model, that both distributes images on the screen and automatically creates visually similar clusters (as a side effect). To refer this tool, you can refer our publications - Image Exploration using Online Feature Extraction and Reranking (ICMR, 2012) and SIR: The Smart Image Retrieval Engine (SISAP, 2012). |
|
Sketch-based Video Browser (or Video Hunter)
(An interactive video retrieval tool for known-item search tasks.)
Sketch-based Video Browser (or Video Hunter) is a new interactive video retrieval tool focusing on known-item search (KIS) tasks. In KIS tasks, users know about some video scene, know it is contained in a collection, but do not know where it is located. Therefore, users have to search/browse the collection with advanced techniques enabling query initialization, result visualization and browsing. Given such support, known-item search is not restricted just to ideal query formulation. The Video Hunter tool has participated in the Video Browser Showdown, winning the competition in 2014 and 2015. In both years 2016 and 2017, the Video Hunter tool achieved the third place. |
|
Web Image Extractor
(Image feature signatures extractor demo implemented in web browser)
A demo which presents a feature extraction method that captures color and texture information from an image and produces adaptive signatures for similarity search models, where distances like SQFD or EMD can be used. The method and its parallel implementation for GPUs is presented in our publication (listed below). We are currently transforming the code, it can be used as OpenCV module. |
SIRET Research Group
Department of Software Engineering
Faculty of Mathematics and Physics
Charles University
Malostranské nám. 25,
118 00 Prague
Czech Republic
| email: | info@siret.cz |
| phone: | +420 95155 4227 |