Bioinformatics & Cheminformatics
 Molpher
(Software tool for exploration of the chemical space)
Molpher
(Software tool for exploration of the chemical space)
Molpher aims to be a scalable and interactive software tool to aid exploration of chemical space, the vast universe containing all possible compounds. Many areas of chemical biology, such as drug discovery, rely heavily on chemical libraries offering compounds usable in the industrial processes. Given a set of molecules with desired characteristics, Molpher explores their common neighbourhood based on structural similarity, as it represents promising part of the chemical space to find new additions into those libraries. In order to decrease the chance of missing interesting parts of the space, Molpher offers the human researcher to observe and interactively alter the exploration process. Generated subspace is expected to be further tested for synthesizability and biological activity by other software tools. Among main features, Molpher offers optimized parallel exploration algorithm, compound logging, dimension-reduced visualization of chemical space and interactive widget-based GUI. Codebase is extensible in terms of additional morphing operators, chemical fingerprints, similarity measures and visualization strategies to allow further experiments. |
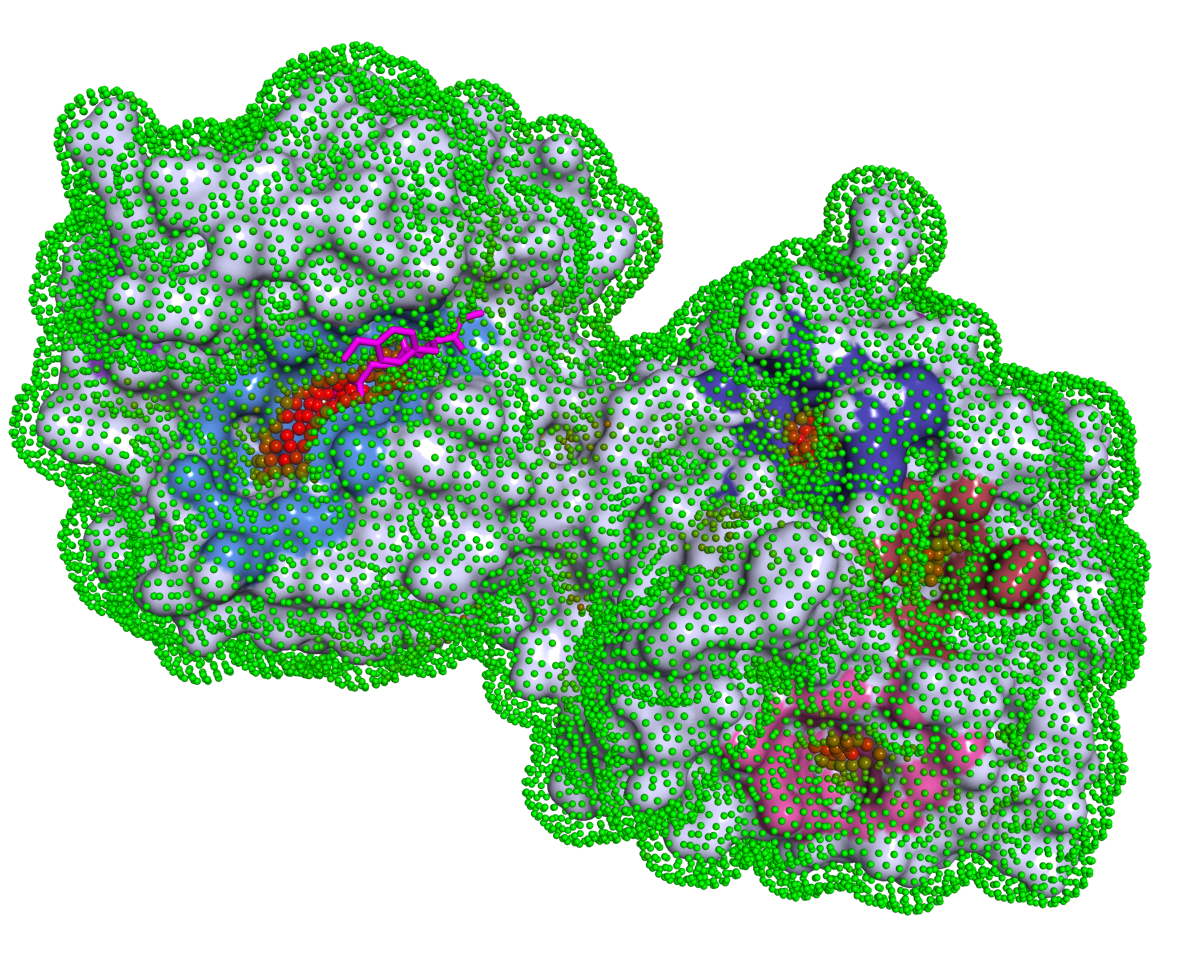 P2Rank
(Protein binding site prediction)
P2Rank
(Protein binding site prediction)
P2Rank is a machine learning based method for prediction of binding sites from protein structure. P2Rank uses Random Forests classifier to infer ligandability of local chemical neighborhoods near the protein surface which are represented by specific near-surface points and described by aggregating physico-chemical features projected on those points from neighboring protein atoms. The points with high predicted ligandability are clustered and ranked to obtain the resulting list of binding site predictions. P2Rank is freely available at https://prankweb.cz/.
|
